MSP-HTPrimer_Manual
TABLE OF CONTENTS
1. What is MSP-HTPrimer?
MSP-HTPrimer is an open source, web-based high-throughput and genome-wide primer design pipeline for bisulfite-based assays (MSP, BSP, PyroSeq, and COBRA) and MSRE-PCR assay and capable of simultaneously processing hundreds to thousands of target sequences. MSP-HTPrimer takes genome-wide annotations of SNPs and repeats into consideration to design primer pairs for higher success rate. MSP-HTPrimer enables hierarchical filtering and visualization of designed primers in UCSC genome browser for efficient selection of assays. MSP-HTPrimer is a user-friendly and standalone tool, which is available within a fully configured Virtual machine. It does not require any installation or configuration except VirtualBox (http://www.virtualbox.com).
MSP-HTPrimer has following unique features:
- Designs primer for BSP, MSP, COBRA and MSRE assays.
- Has no limitation for number and size of target requences for primer design in parellel.
- Design BSP, PyroSeq, COBRA and MSP primers on both forward and reverse strand of target sequence.
- Takes common SNPs and repeat regions into account in primer design.
- Visualizes primer pairs in UCSC genome browsers.
- Has intutive and user-friend web interface especially designed for non-expert users.
- Allows automated primer ranking and selection based on user defind crieria.
2. System requirements
Virtual box from https://www.virtualbox.org/
Operating system
Linux
Mac OSX 10.6 or later
Windows PC
3. How to obtain MSP-HTPrimer?
MSP-HTPrimer is a web-based standalone tool. We provide source code in two version of MSP-HTPrimer for Linux, and Macintosh operating systems. For Windows PC users we provide a fully configured Virtual Machine (VM can be used on any operating system). Along with source code and virtual machine we provide test data and extensive user manual for step-by-step get and run MSP-HTPrimer for expert and non-expert users.
3.1 Download MSP-HTPrimer Virtual Machine
Fully configured Virtual box and can be downloaded from https://sourceforge.net/p/msp-htprimer/wiki/Virtual_Machine and can be easily run on any operating system. With Virtual machine no installation and configuration required.
3.1.1 How to use MSP-HTPrimer Virtual Machine?
Once virtual machine of MSP-HTPrimer is obtained then follow these steps to run the MSP-HTPrimer tool.
Step1.1: Download and install the Virtual Box (version 5.1.2) from http://www.oracle.com/technetwork/server-storage/virtualbox/downloads/index.html#vbox
Step1.2: After installation of Virtual Box download and install the Virtual Box Extension Pack (version 5.1.2) from http://download.virtualbox.org/virtualbox/5.1.2/Oracle_VM_VirtualBox_Extension_Pack-5.1.2.vbox-extpack
Step2: Import MSP-HTPrimer Virtual Machine file into Virtual Box
Step3: Login into MSP-HTPrimer Virtual machine with username = testuser and password = testuser
Step4: Open Firefox or any other web browser and open the query page of MSP-HTPrimer with http://localhost/msp-htprimer
Step5: Run the MSP-HTPrimer with the test data sets.
Step6: For new data analysis with MSP-HTPrimer, prepare the Target file and run the primer design.
3.2 Download MSP-HTPrimer source code
To use MSP-HTPrimer outside VirtualBox on local server, user can download the latest version of MSP-HTPrimer source code (1) for Linux computers from https://sourceforge.net/projects/msp-htprimer/files/Linux and 2) for Macintosh OSX computers form https://sourceforge.net/projects/msp-htprimer/files/MacOS. Once source code is downloaded configure MSP-HTPrimer web interface by following the instructions given in README file (available from above URL).
4. MSP-HTPrimer web interface description
4.1 MSP-HTPrimer query page
User can run primer design with MSP-HTPrimer by providing input options and files from query page as shown in Figure 1.
Following input parameters and input files are required.
Input 1:
Genome information parameters are required to download genome fasta sequence and annotation files (RefSeq gene, common SNPs, CpG island and known repeat elements) from UCSC genome browser (http://genome.ucsc.edu/index.html)
1) Select genome name from first drop down menu (Human or Mouse). The default genome is Human.
2) Select genome assembly version from second drop down menu (default genome assembly is hg19)
3) Select the dbSNP build to download the corresponding common SNPs from UCSC genome browser. Default is 142 for Human, genome assembly hg19.
**Input 2: **
Upload a target file in BED format. One file for each target region. This file consists of 4 columns 1) chromosome, 2) start position, 3) end position and 4) target ID. User can give any number of target region in a single run.
Input 3:
Third input is the Primer3 input parameters for primer design. This file can be modified as per the requirement and upload. This is an optional input, if user does not provide then MSP-HTPrimer uses the default setting of Primer3.
Input 4:
This input is only required fo BSP-COBRA and MSRE primer design, user can either enter type-II enzyme is the input box (one enzyme per line) or alternatively can upload a text file which contains one enzyme per line.
Input 5:
To provide flexibility in primer design process, MSP-HTPrimer provides some useful input options for optimized and speific primer design and selection. Under these parameters user can define
1. Maximum primer pairs to return for each target region. Default 10
2. Product size: Minimum, Optimum and Maximum. Default 150, 250 and 320 respectively.
3. Primer annealing temperature. Default 52, 60 and 65 for Minimum, Optimum and Maximum temperature respectively.
4. Primer size: Minimum, Optimum and Maximum. Default 22, 28 and 36 bp respectively.
5. Product CpGs: Minimum number of CpG in PCR product. Default 4.
6. CpG in primer: Minimum number of CpG in primer pairs. Default 1.
7. Primer non-CpG 'C's: Minimum number of non CpG C's in primers (especially for BSP and COBRA). Default 4.
8. Primer Poly X: Maximum number of consicutive non T's mononucleotide. Default 5.
9. Primer Poly T: Maximum number of consicutive T's. Default 8.
** Parameters for Pyrosequencing primers**
1. Window size: Give a window size to design sequencing primer in a sliding window approach over whole amplicon. Default 100 bp.
2. Step size: Give a overlap size in two consicutive windows. This parameter is useful for overlapping sequencing primer design. Default 20 bp.
3. PyroSeq primer size: Minimum, Optimum and Maximum. Default 15, 18 and 25 bp respectively.
4. PyroSeq primer Tm: Default 25, 28 and 30 for Minimum, Optimum and Maximum temperature respectively.
5. PyroSeq product CpG: Minimum number of CpG in PCR product. Default 0.
6. CpG in PyroSeq primer: Minimum number of CpG in primer pairs. Default 0.
7. PyroSeq primer non-CpG 'C's: Minimum number of non CpG C's in primers. Default 4.
8. PyroSeq primer Poly X: Maximum number of consicutive non T's mononucleotide. Default 5.
9. PyroSeq primer Poly T: Maximum number of consicutive T's. Default 8.
** Parameters for Hyb. probe design**
1. Hyb Proble Size : Default size 18, 20 and 27 for Minimum, Optimum and Maximum respectively.
2. Hyb Probe Tm : Default 52, 60 and 65 for Minimum, Optimum and Maximum temperature respectively.
3. Hyb Probe GC% : Default 20, 50 and 80 for Minimum, Optimum and Maximum GC content.
Parameters for MSP primers
1. 3'CpG constraint: Position of CpG at the primer's 3' end. Default 3.
2. Max Tm difference: Maximum Tm difference between Methylated and Unmethylated primer. Default 5 degree.
3. Product length difference: methylated and unmethylated product length difference. Minimum and Maximum range can be defined. Defult 0.
Input 6:
This is unique feature of MSP-HTPrimer to provide primer selection quality matrix for final primer pair selection, which helps to reduce the post selection process. User can define various filtering criteria for each output column of the MSRE-HTPrimer and then tool automatically selection primers from the whole output. This is optional input.

Figure 1: shows MSP-HTPrimer query page to define all parameters and upload input files for primer design
4.2 MSP-HTPrimer result page
4.2.1 MSP-HTPrimer result page for MSP primer design
MSP-HTPrimer results all primer pairs in a summary table, which is available in HTML to display (as shown in Figure 2) and in TXT and HTML format to download. Moreover, MSP-HTPrimer has seamlessly integrated the UCSC genome browser visualization in the result page. All resulting primer pairs of a target region is visualized in UCSC genome browser as shown in Figure 2. The ampilcon and its primer pairs for methylated target are dispalyed in maroon color and for unmethylated target displayed in blue color.

Figure 2: shows MSP primers designed by MSP-HTPrimer tool. Primer pair output summary table (top panel) (http://localhost/msp-htprimer), and visualization of primer pairs in UCSC genome browser within the MSP-HTPrimer interface (bottom panel). Go to UCSC genome browser
4.2.2 MSP-HTPrimer result page for PYROSEQ sequencing primer design
Figure 3 shows the Pyrosequencing primer design result produced by MSP-HTPrimer. The primer summary table contains Ts_Id, Fp_Seq, Rp_Seq, Amp_Id, Amp_Bed, PyroSeq_Primer_Seq, and UCSC_Genome_Browser link. A detailed output summary table can be downloaded in TXT and HTML format.
All resulting primer pairs of a target region is visualized in UCSC genome browser as shown in Figure 3. In UCSC genome browser The Amplicon and BSP amplification primer pairs are displayed in maroon color. The Pyrosequencing primers are displayed in blue color.

Figure 3: shows Pyrosequencing primer designed by MSP-HTPrimer tool. Primer pair output summary table (top panel) (http://localhost/msp-htprimer), and visualization of primer pairs in UCSC genome browser within the MSP-HTPrimer interface (bottom panel). Go to UCSC genome browser
4.3 Run MSP-HTPrimer with test input
To validate the installation of the MSP-HTPrimer pipeline it can be run with a small test data set. The test data set for BSP, BCP-COBRA, MSP-PCR andMSRE-PCR can be obtained from https://sourceforge.net/projects/msp-htprimer/files/test_data.zip
and run the following command to uncompress the file:
unzip test_data.zip
Note that after uncompressing the .zip file, a new folder will be created named test_data. Now upload these files on the MSP-HTPrimer query page (http://localhost/msp-htprimer) and run the primer design.
5. MSP-HTPrimer inputs description
MSP-HTPrimer requires four input files:
5.1 Target BED file
This file contains the genomic coordinates for all target sequences (one line for each target sequence). It consists of four tab-delimited columns: 1) chromosome, 2) start coordinate, 2) end coordinate and 4) a unique ID for each target region as shown in Table below.
chr2 241454334 241457334 Target1
chr3 10155818 10158818 Target2
chr5 118813546 118816546 Target3
chr5 148183848 148186848 Target4
chr5 112098954 112101954 Target5
chr15 89059082 89062082 Target6
chr19 1154297 1157297 Target7
chrY 25386895 25389895 Target8
5.2 Primer3 parameter file
This text file contains the parameters and values for the Primer3 tool. It is optional and if not provided, MSP-HTPrimer will use default Primer3 parameters as shown below:
PRIMER_TASK=generic
PRIMER_MISPRIMING_LIBRARY=
PRIMER_MIN_TM=65.0
PRIMER_OPT_TM=70.0
PRIMER_MAX_TM=75.0
PRIMER_MIN_GC=20.0
PRIMER_MAX_GC=100.0
PRIMER_NUM_RETURN=5000
PRIMER_MIN_SIZE=16
PRIMER_OPT_SIZE=21
PRIMER_MAX_SIZE=30
PRIMER_PRODUCT_SIZE_RANGE=50-150
SEQUENCE_ID=TS001
SEQUENCE_TEMPLATE=
PRIMER_PICK_LEFT_PRIMER=1
PRIMER_PICK_RIGHT_PRIMER=1
PRIMER_PICK_INTERNAL_OLIGO=1
PRIMER_PICK_ANYWAY=1
PRIMER_THERMODYNAMIC_OLIGO_ALIGNMENT=0
PRIMER_THERMODYNAMIC_TEMPLATE_ALIGNMENT=0
5.3 Restriction enzyme file
This input file is only required for BCP-COBRA and MSRE-PCR primer design. Each line contains an enzyme name as per nomenclature and multiple enzymes are allowed in a single run as shown below:
MSRE enzymes
HpaII
Hin6I
AciI
HpyCH4IV
COBRA enzymes
BstUI
TaqI
5.4 Custom primer selection quality matrix
MSP-HTPrimer supports further selection of primer pairs based on user defined selection criteria. A custom quality-filtering matrix can be provided as input file. As shown in Table below, the user can define a set of selection criteria and rank them using a scale of 1-10. MSP-HTPrimer assigns these ranks to the primer pairs for all target sequences. If this input is not provided then primer pairs are returned based on Primer3 ranking. MSP-HTPrimer supports mathematical operators, including “>”, “<”, “>=”, “<=” and “-“. Any column header of the MSP-HTPrimer output file can be used as parameter. The primer quality level represents the rank associated with each of the output parameters in its respective row.
Table: Custom quality filter matrix with ten quality levels ranking the designed primer independent of the primer3 level, but dependent on amplicon size, amount of cutsites and gene distance.

All headers consist of two major parts, origin (Fp=forward primer, Lp=left primer, Rp=reverse primer/right primer, Amp=amplicon, Hyb=hybridization oligo) and short description. Primer_Quality_Level=user defined rank; Tm=melting temperature of origin; Gc_%=GC percentage in DNA sequence of origin; Any_Compl=stability of any basepairing of origin to itself; 3'_Compl= stability of any basepairing of the 3' end of the origin to itself; Size=size of origin in basepairs (Bp); Repeat_In_Bp=allowed Bp of repeats in origin; Snp_Pos_From_3'=distance of closest SNP position to 3’end inside the origin sequence in basepairs; Amp_Sum_Cutsites_Primer=amount of cutsites in FP and RP; Amp_Sum_Cutsites_Between_Primers=amount of cutsites in amplicon except for FP and RP.
6. How to use MSP-HTPrimer?
To use MSP-HTPrimer for primer design user
Open web browser and type the following url into browser (http://localhost/msp-htprimer) and upload required inputs files, change default parameters if required and run the primer design.
7. Performance evaluation
MSP-HTPrimer is a high-throughput primer design pipeline and can design primers for ten to several hundred target regions simultaneously. To evaluate the performance of MSP-HTPrimer, from Human RefSeq genes (Hg38), we have randomly selected 500 target sequences of 1 kb length (±500 bp to the Transcription Start Site) which falls within CpG island regions. The benchmarking was performed on a Linux server (Ubuntu 14.0.4 LTS with 8 CPU, 16 GB RAM). Execution times were measured for all four methods BSP-PCR (black), MSP-PCR (blue), COBRA-PCR (green), and MSRE-PCR (red). All benchmark measurements have been performed using the Primer3 version 2.3.6 with a maximum of 200 primer pairs to return per target sequence. All execution times were measured in seconds. As shown in Figure 4, MSP-HTPrimer is very fast and efficient to design specific primer pairs for hundreds of target regions. As shown design for 100 MSP-PCR assays is conducted in nearly 1755 seconds (~29min) computing-time to run the entire steps according the pipeline. For the same dataset MSP-PCR design takes more time than other methods (e.g. BSP: 608 seconds, COBRA: 731 seconds, and MSRE: 216 seconds for designing 100 assays), which is due to the two pair of primer designs (methylated and unmethlated target sequence), and checking the compatibility of both primers (methylated and unmethylated) and their PCR products (Figure 4).
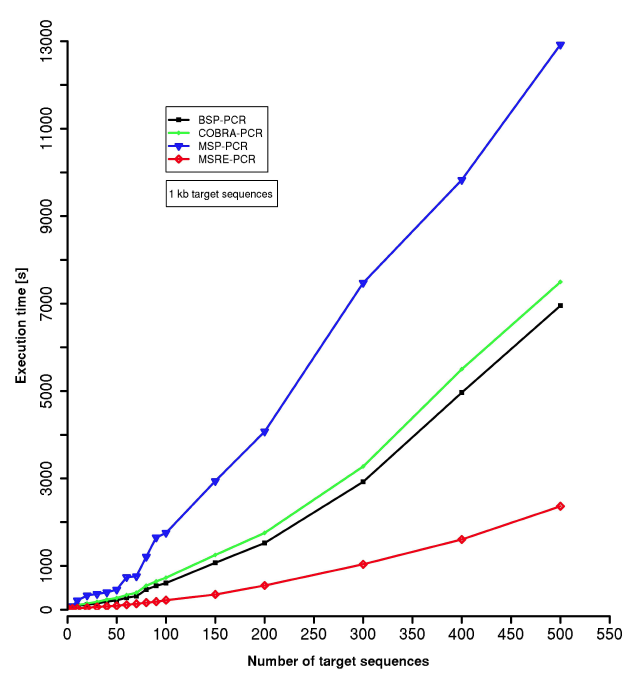
Figure 4:Evaluation of MSP-HTPrimer execution for BSP (black line), MSP (blue line), COBRA (green line) and MSRE (red line); considering number of 1kb long target sequences.
8. Contact Information
Ram Vinay Pandey
ramvinay.pandey@gmail.com

