Usage
Biosig libraries
The libraries are not useful on there own, but provided the core functions for accessing and manipulating biomedical signal data in various formats. Internally, a common programming interface to all supported data formats is provided. It makes also use of an automated file formation identification based on magic keys.
Biosig provides the following libraries:
- libbiosig: supports reading of about 50 differernt data formats, and writing of about 10 different formats. The complete list is maintained at http://pub.ist.ac.at/~schloegl/biosig/TESTED
- libgdf: provides the same API than libbiosig, but supports only GDF file format. This might be useful in order to reduce the potential attack surface, has much lower prerequisites.
- libphysicalunits: provides conversion funtions for encoding and decoding physical units according to ISO 11073-10101. This encoding is used in GDF and internally in libbiosig; libbiosig provides these as well, but they might be usefu on their own as well.
- libgdftime (is part of libbiosig): Internally, the format for encoding date and time uses a 64bit data type, where the first 32 bits are used to encode the number of days, and the sencond 32bits are used to encode fractions of a day. This enables a time resolution of about 20 us, conversion functions are provided here.
Biosig-tools
There are a number of command line tools for data conversion. Most importantly,
* save2gdf : can read about 50 dataformats, and as the name suggests, it converts it to the GDF format. Besides that in can also write to a number of other (~10) formats, some combinations might not be well tested. It can also produced JSON, ASCII output of the metainformation (header, events/marker/annotation, etc.)
-
save2aecg: uses save2gdf, and converts short term ECG data into the XML-based HL7v3/annotated ECG format.
-
save2scp: uses save2gdf, and converts short term ECG data into EN1064 (a.k.a. ISO 11073-91064) SCP-ECG file format .
-
bin2rec, rec2bin: use save2gdf, for converting biosignal data into multiple ASCII files (1 header file, and one file for each channel), and for the reverse conversion
-
biosig_fhir: is a tool that can convert the data into a binary format confirming to the binary template format of HL7/FHIR using XML or JSON format.
-
physicalunits :is a command line tool for encoding and decoding physical units .
mexBiosig
provides the mex-functions mexSLOAD, mexSOPEN, mexSSAVE, physicalunits . All data formats supported by libbiosig are also supported by mexBiosig. Please note, a number file formats are supported by libbiosig and mexBiosig, which are not supported by biosig4octmat ( see (9) in
http://pub.ist.ac.at/~schloegl/biosig/TESTED ). However, biosig4octmat can make use of mexBiosig when available.
Biosig for Octave and Matlab (a.k.a. biosig4octmat)
is a large set of m-file scripts and functions for biomedical signal processing. The functions are grouped into several subdirectories (t200 for file access, t250 for preprocessing, t300 for signal processing and feature extraction, t400 for classifification, t500 for visualizaiton, etc.) . It contains numerous useful functions for EEG analysis, EEG-based Brain-computer-interface, processing of EEG artifacts, sleep classification, Detection of action potentials, sharp-wave ripples, etc. An extensive function index can be found here http://biosig.sourceforge.net/help/index.html .
Biosig for Python
provides currently the functions
- biosig.header
- biosig.data
- biosig.error
which read the metainformation (header, annotations/markers/event), the data samples, and a function for error checking. A complete example is available here https://sourceforge.net/p/biosig/code/ci/master/tree/biosig4c++/python/demo2.py?format=raw .
A screenshot of running the demo under Cygwin is shown here 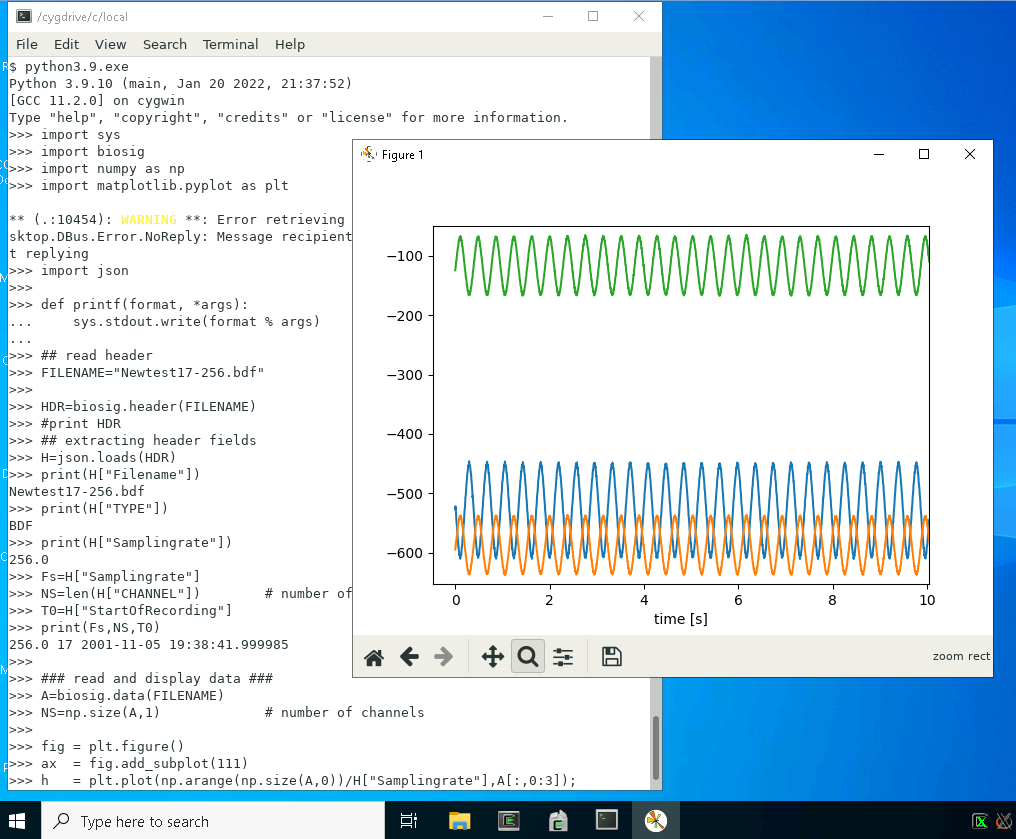
Biosig on Cygwin
... is now available here and here. This is currently the recommended way to use Biosig for Python on Windows (an alternative might be WSL but this is not tested).
"Biosig for Python" has already been possible for users of *nix operating sytems (including GNU/Linux, MacOSX and other *nix). Using Cygwin, this now also available to users of the Windows OS.
Most likely when using Biosig in Python, you will might want displaying some data with matplotlib. In order to do so, you will need also Cygwin/X. Above, a screenshot is shown about loading and displaying some demo data, running Python3.9.exe on a mate terminal under Cygwin/X.
Best regards.
Alois

