Here's a very simple model which infers a population sd and effect size from one-sample data with known population mean. The effect size is the deviation of the sample from this population mean, standardized to SD's. I've used JZS-priors.
The problem is that "version 1" (with a small measurement error) somehow has very bad mixing while "version 2" (without measurement error) does not. If anyone can explain why this arises in version 1 but not version 2 - and maybe even fix it - that would be much appreciated.
# Priors on population parameters
pop.pr ~ dgamma(0.00001,0.00001)# approximation to Jeffrey's prior on variance
pop.sd <-sqrt(1/ pop.pr)# convert to SD# Priors on (population) effect size
effect.d ~ dt(0,1,1)# cauchy prior on standardized effect size (Cohen's d)
effect.abs <- effect.d * pop.sd # convert to absolute effect sizefor(i in1:length(x1)){# Version with Bad mixing: with measurement error (SD=1)
ability[i,1]~ dnorm(100, pop.pr)
x2[i]~ dnorm(ability[i,1]+ effect.abs, pow(1,-2))#Version with relatively good mixing: without measurement error#x2[i] ~ dnorm(100 + effect.abs, pop.pr)}
I ran it on simulated data with mu=100, sd=15, d=2 (large effect) and n=100. With measurement error, there's a clear inverse relationship between estimated population sd and Cohen's d.
Simulate data in R:
We're just going to use x2 for this simple simulation. It's a measurement after an intervention.
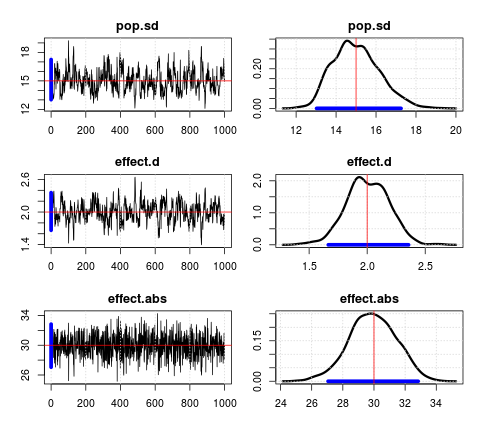
All looks pretty good. Not perfect mixing on effect size and sd, but good enough.
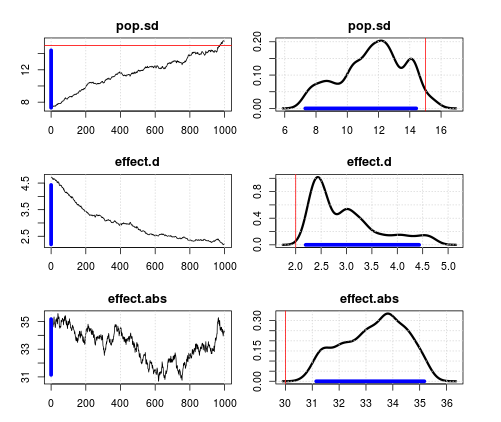
Result with measurement error
Horrible mixing. It is even worse if the measurement error is decreased. That makes no sense to me since that should approximate the results from above.
This is a weakness of sampling node-by-node. The sampling efficiency depends on how you parameterize your model.
Your example would work fine if you were using the glm module and if effect.abs had a normal distribution. Then effect.abs would be updated in a block with ability[,1] and you would get good mixing, but the glm model cannot (yet) handle variables with a t distribution so this opportunity is missed.
If you would like to refer to this comment somewhere else in this project, copy and paste the following link:
Thanks, Martyn! That solved it for this particular problem, but mixing remained a problem when I tried to extend the model to repeated measures (x2 = x1 + effect). The whole purpose was to put the prior on Cohen's d rather than the absolute effect size, in order to do a JZS-prior. I figured out a solution which is mathematically equivalent to the script above but yields good mixing.
Instead of
# Priors on (population) effect size
effect.d ~ dt(0,1,1)# cauchy prior on standardized effect size (Cohen's d)
effect.abs <- effect.d * pop.sd # convert to absolute effect size
I did
# Priors on (population) effect size
effect.abs ~ dt(0, pop.sd,1)# cauchy prior on cohen's d of this effect
effect.d <- effect.abs / pop.sd
Exactly why this works, I'm not sure. But the new model is simpler (cohen's d is just computed post-hoc), which is a nice thing.
Best,
Jonas
If you would like to refer to this comment somewhere else in this project, copy and paste the following link:
Here's a very simple model which infers a population sd and effect size from one-sample data with known population mean. The effect size is the deviation of the sample from this population mean, standardized to SD's. I've used JZS-priors.
The problem is that "version 1" (with a small measurement error) somehow has very bad mixing while "version 2" (without measurement error) does not. If anyone can explain why this arises in version 1 but not version 2 - and maybe even fix it - that would be much appreciated.
I ran it on simulated data with mu=100, sd=15, d=2 (large effect) and n=100. With measurement error, there's a clear inverse relationship between estimated population sd and Cohen's d.
Simulate data in R:
We're just going to use x2 for this simple simulation. It's a measurement after an intervention.
Results without measurement error
All looks pretty good. Not perfect mixing on effect size and sd, but good enough.
Result with measurement error
Horrible mixing. It is even worse if the measurement error is decreased. That makes no sense to me since that should approximate the results from above.
By the way, my earlier post on another simple mixing problem remains unanswered: https://sourceforge.net/p/mcmc-jags/discussion/610037/thread/aae85a9e/
Thanks for any help!
Last edit: Jonas Lindeløv 2014-04-03
This is a parameterization problem. If you rewrite the model in the form below you get much better mixing:
This is a weakness of sampling node-by-node. The sampling efficiency depends on how you parameterize your model.
Your example would work fine if you were using the glm module and if
effect.abshad a normal distribution. Theneffect.abswould be updated in a block withability[,1]and you would get good mixing, but the glm model cannot (yet) handle variables with a t distribution so this opportunity is missed.Thanks, Martyn! That solved it for this particular problem, but mixing remained a problem when I tried to extend the model to repeated measures (x2 = x1 + effect). The whole purpose was to put the prior on Cohen's d rather than the absolute effect size, in order to do a JZS-prior. I figured out a solution which is mathematically equivalent to the script above but yields good mixing.
Instead of
I did
Exactly why this works, I'm not sure. But the new model is simpler (cohen's d is just computed post-hoc), which is a nice thing.
Best,
Jonas