Home
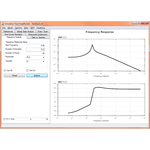
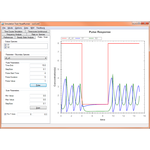
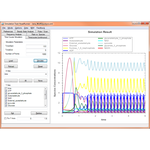
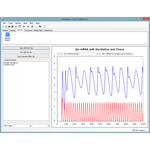
This project hosts the 'Simulation Tool', an extensible, interactive environment for carrying out simulation experiments of SBML models. This project can use RoadRunner or SBW enabled simulators to carry out simulations.
For now if you are interested in the project, you can download a windows version included with the Systems Biology Workbench here:
http://sbw.kgi.edu/fbergman/files/latest/SetupSBW.exe
Instructions for other platforms and virtual machines
If you'd like to develop for the SBW simulation tool have a look at the source here:
http://sbwsimtool.svn.sourceforge.net/viewvc/sbwsimtool
The simulation tool directly supports SED-ML L1V1, and allows you to execute the simulations directly, with support for remote models (even in BioModels.net) and the SED-ML archive.