Home
BAIT - A Comprehensive Analysis Tool for Strand-Seq Data
Publication:
Please reference: Hills et al., 2013. (Actual publication info to follow).
Original Strand-seq paper published in Nature Methods by Falconer et al., 2012
Tutorials:
What is Strand-seq and how does it work?
Tutorial for strand inheritance studies
Tutorial for sister chromatid exchange studies
Tutorial for identifying genomic rearrangements
Tutorial for localization of orphan fragments
Tutorial for building early stage genomes
Dependencies:
- Requires samtools set in PATH (Li H. et al. (2009) Bioinformatics, 25:2078-9) or location set using -3 option
- Requires bamToBed set in PATH (Quinlan A. and Hall I. (2010) Bioinformatics, 26:841–2) or location set using -4 option
- Requires DNAcopy library in R (Venkatraman ES & Olshen AB (2007) Bioinformatics, 23:657-663)
- Requires GenomicRanges library in R (Laurence et al. (2013) PLoS Computational Biology, 9)
- Requires changepoint library in R
- BAIT will not execute if BAM header is absent
- BAIT expects a 6nt index within bam files, or a 3 digit miSeq index (use -m option)
- BAIT expects to see a header with SN: and LN: lines to build the chromosome numbers and lengths
- BAIT expects a SP: line to indicate the species. If this line is not present in the header, BAIT will prompt for this information (can be set using -7 and -8 options)
- Current supported species include: Mus_musuclus, Homo_sapiens, Caenorhabditis_elegans & Saccharomyces_cerevisiae. Other organisms will still generate plots, but without exclusion regions or optimally adjusted window size (can be set using -w option)
Introduction:
BAIT (Bioinformatic Analysis of Inherited Templates) is a software package for comprehensive screening of Strand-seq data. Bam files are sequentially parsed through a shell script and processed for a variety of downstream applications. BAIT allows end-users to identify strand inheritance, sister chromatid exchanges (SCEs), translocations, genome errors, and can aid in the building and finishing of genomes.
For a more complete synopsis, please refer to Falconer et al., Nature Methods (2012), for a summary of Strand-seq and its applications, and Hills et al., REF TO FOLLOW (2013), for a summary of the program itself. Strand-seq data typically consists of multiple sequence libraries, where each library is derived from a single cell. Briefly, stands are distinguished based on the incorporation of the thymidine analogue BrdU, resulting in only template strands remaining within the libraries. Strands are designated as Watson (W) or Crick (C) depending on strand directionality, and so in diploid cells, each chromosome can have a homozygous template state (WW or CC) or a heterozygous state (WC), at a ratio of 1:1:2 respectively. By maintaining directionality, it is possible to assess which templates were inherited for each chromosome and locations where those inheritance patterns change are sites where SCE have occurred (or, if recurrent, are potentially regions of gross genomic alterations). Using a large Strand-seq data set across multiple libraries it is also possible to aid in finishing and building genomes. Mis-oriented regions in genome builds can be readily identified as a complete switch in template state at the same location in every library. Unlocalized scaffolds can be located by comparing the template state of the scaffold to the template state of all the chromosomes. For example, if an unlocalized scaffold is WW in a library, it will map to a chromosome in that library that is also WW. Comparisons across large datasets allow a concordance calculation that establishes how well template strands of a particular scaffold coincide with each chromosome. Finally, extending these analyses further, we can compare contig-level genome builds and assess which contigs share the same strand inheritance pattern across all libraries, and infer that if this pattern is coincident, then these contigs must be on the same chromosome and close together.
Pipeline:
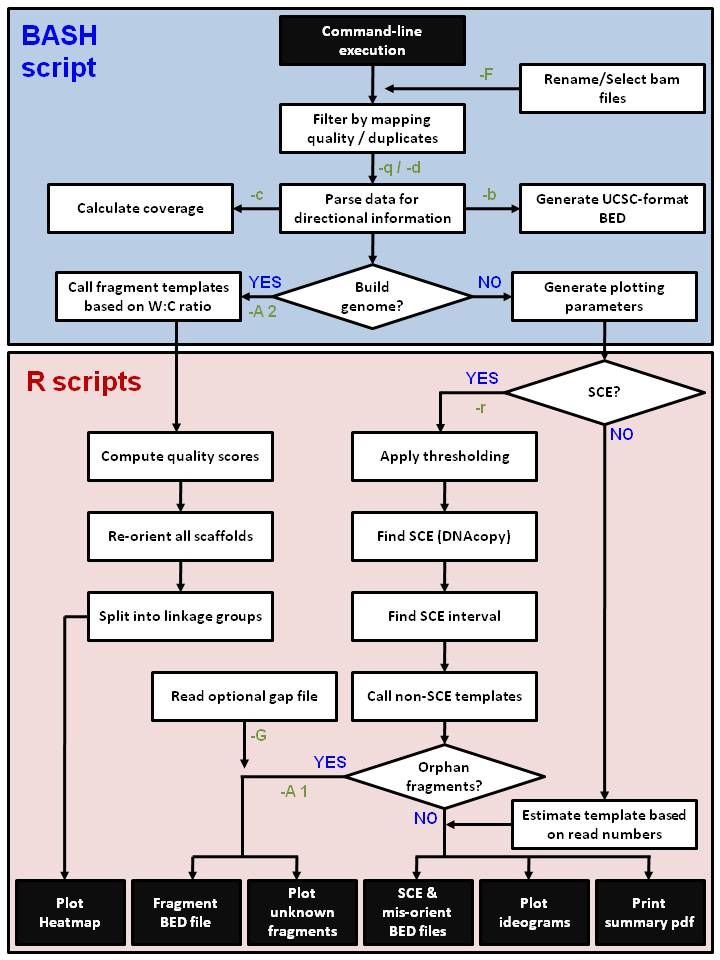
Installation:
Unzip BAIT and place it in a directory of your choice. for example:
tar -jcvf BAIT_v1.0.tar.bz /usr/local/bin/BAIT_v1.0/
Copy the BAIT executable ('BAIT') to a directory in your PATH. For example, to save to the /usr/local/bin folder, type:
sudo ln -s /usr/local/bin/BAIT
Set an environment variable (e.g. in your .bashrc) named BAITPATH to the directory in (1). For example, if the BAIT folder is present in /usr/local/bin/:
(a) access your bashrc file by opening a text editor, navigating to your home directory and typing ".bashrc".
(b) add the following to the end of the .bashrc file:
export BAITPATH="/usr/local/bin/BAIT_v1.0"
(c) save this file.
Or
(a) open a terminal.
(b) type the following:
(cat ~/.bashrc; echo "export BAITPATH="/usr/local/bin/BAIT_v1.0"") >> ~/.bashrc
(c) the .bashrc file should now contain the additional line, and BAIT should run correctly
You should now be ready to run BAIT.
Usage:
BAIT has several user options to perform a variety of Strand-seq analysis. The options can be found using BAIT.sh -h to bring up the help menu. The following descriptions are intended to provide more detail for each option:
STANDARD ANALYSIS OPTIONS
-i PATH
INPUT. Specifies the input folder (default is current folder). This option is used to direct BAIT to the location of the Strand-seq bam files. The default folder is the folder in which BAIT is being run (the current directory), but if data are stored in a different directory to the current folder, it
is possible to designate a PATH to where BAIT can find these files.
-o STR
OUTPUT. Output file name (default 'BAIT'). This option will create a name for all output data and folders. The default name is 'BAIT'. A warning will execute if a folder already exists in the
working directory with the same name. An option to either delete these folders and overwrite, or to rename the output files will be given.
-a
ARROW. Plots arrowhead on ideograms denoting detected SCE events and mis-orientations. Mis-orientations are plotted in red, while SCEs are plotted in black. Can only be used in conjunction with the -r option.
-c
COVERAGE. Adds % coverage information (requires -g option). This option will run a pileup for each sample, remove all N bases, then calculate the percentage coverage by dividing this value by
the number of bases present in the genome. In order for pileup to execute, samples need to be aligned to a genome which must be set in the PATH using the -g option.
-d
DUPLICATES. Turns off duplicate removal; not recommended. If samples have already had duplicate reads removed, or to speed up run times, files can be processed without prior removal of duplicates.
BAIT assumes paired end reads, so for single end, run with the -d option.
-p
PHASING. Performs haplotype phasing (requires -g option). Currently in beta. This option significantly increases run time. Haplotype phasing is achieved by taking only CC or WW chromosomes
from each library. Initial establishment of heterozygous SNVs is achieved either by inputting a non-Strand-seq, whole genome sequence bam file at the prompt, or by automated merging of all libraries and using this as a SNV reference key. All heterozygous regions defined in this reference key are scanned for each library, and SNPs on the same chromosome are systematically phased. Requires running a pileup, so samples need to be aligned to a genome which must be set in the PATH using the -g option.
-q INT
QUALITY. Minimum mapping quality for analysis (default 10).
-b
BED OUTPUT. Generates UCSC-formatted BED files. This option will create BED-formatted Strand-seq files in a format acceptable for UCSC upload. Reads mapping to Watson strands coloured are blue and reads mapping to Crick are coloured orange, similar to the ideogram plots.
-B PATH
BED FILE. Allows plotting of optional BED files onto ideograms. The file can be in any BED format but BAIT only looks at the first 4 columns. The forth column should either be blank or specify a colour compatible with R. If the forth column is empty, BAIT will automatically designate a plotting colour (default is red, but plotting colours can also be set in the command line but invoking the -C option.
-C STR
COLOUR. Specifies colour of plotted optional BED file (default 'red') if using the -B option. Can be any R-format colour name or colour reference number (full list found HERE).
-G PATH
GAP FILE. Allows plotting of optional BED-format GAP file onto ideograms. Generates white spaces for gaps onto the ideogram plots. Also read in to determine most-likely intervals for unplaced fragments if -u option is invoked.
-g PATH
GENOME. Specifies the location of an indexed fastq-format genome to allow -p and -c options.
-F PATH
FILE. Read in file for sample info. File should be in csv format with the index from the file name in column 1 and the name to be used for the plotting in column 2. When -F is invoked, only those samples
that are present in the supplied file will be analyzed by BAIT. All other bam files in the working directory will be ignored.
-r
RECOMBINATION. Analyzes sister chromatid exchanges for each library. This function executes the DNAcopy circular binary segmentation algorithm, iterating down to the narrowest SCE interval. It then attempts to refine the interval further by looking for the first instance of a change in state. This function must be called for -a, -u , and -p options to be executed.
-k
KEEP. This option retains all intermediate files. Resolves a conflict for running BAIT simultaneously from two input directories. As standard, intermediate data is stored by default to /tmp/BAIT, and will be removed once the program has finished. Using this option will create a folder in your working directory called 'KEPT_FILES_BAIT/'. Option is also useful if the Linux set up does not allow you to access /tmp/, or if intermediate files from large datasets need to be retained for later analysis.
-A INT
ASSEMBLE. This option is used for assembling genomes, and has three different analyssi types. Using -A 1 is for locating orphan scaffolds in complete genomes, and when invoked, BAIT locates unplaced contigs in complete-build genomes. It will compare all unplaced/unknown scaffolds from the bam file to strand inheritance from each chromosome to determine a likely location. This option produces a pdf plot for each unplaced scaffold, and generates csv file of unknown chromosome fragments. The option will not execute if there are no unplaced fragments in the header of the bam file. Some sequencing centres do not include unlocalized and unplaced scaffolds when performing alignment. Unplaced fragments are determined by the presence of an '_', a '-' or a '.' in the SN: line of the bam header. If this option is used with -G, the program will also produce a csv file of all gap locations that are coincident with the predicted locations of the unplaced scaffolds. Requires the -r option to be invoked. Using -A 2 is for building scaffold/chromosome-stage genomes from contigs. It will compare all contigs with each other and generate a heat map that can be used to look for concordance between contigs. It orients contigs with respect to each other and executes a TSP algorithm which attempts to sort the contigs into a correct order. Output is given as a heat map pdf, and a csv file containing the optimum order of contigs. Using -A 3 is for complete genomes with >100 unplaced scaffolds. When invoked, BAIT splits chromosomes based on SCE locations and plots these and the scaffolds on a heat map, locating all scaffolds to interals along each chromosome.
-v
VERBOSE. Prints verbose output, generating messages at each stage of the analysis.
-h
HELP. Prints help page.
TROUBLESHOOTING OPTIONS
In cases where BAIT is crashing or is unable to nd certain information, the following options can be used to troubleshoot errors or alter output.
-1 PATH
BAIT PATH. Defines path to the BAIT folder if not already set in PATH. This option is useful if the .bashrc file has not been changed as described in the installation instructions.
-2 PATH
R PATH. Defines path to Rscript if not already set in PATH. This option is useful if you do not have an administrator account and cannot execute R from the PATH.
-3 PATH
SAM PATH. Defines path to samtools if not already set in PATH. This option is useful if you do not have an administrator account and cannot execute samtools from the PATH.
-4 PATH
BED PATH. Defines path to BamToBed if BEDtools is not set in PATH. This option is useful if you do not have an administrator account and cannot execute BEDtools from the PATH.
-5 PATH
EXCLUSION FILE. User-defined exclusion file. Certain regions are prone to cluster low-complexity reads which will always map to both strands. These regions can be specifically excluded by uploading a BED file where each line contains a region to be excluded. Build-in exclusion regions are provided for the two latest builds of mouse and humans (more organisms to follow).
-6 STR
WINDOW SIZE. User-defined window size. This alters the size of the binning window for BAIT ideogram plotting. The default plotting window size is 200000 bp but for organisms with small genomes, this value may be too large. A good rule of thumb is to generate at least 100 bins per chromosome (ie default is good for organisms where chromosome length > 20Mb). For low coverage libraries, especially those derived from MiSeq (or other desktop sequencers), the default window size might be too small, as there will be few reads per bin. In these instances, increasing window size to 1000000 is recommended.
-7 STR
ORGANISM. Specifies organism/species. This option will override the SP: line found in the header of the bam file. If there is no SP: line BAIT will prompt for an organism unless this option is selected. Therefore, if BAIT is integrated into a pipeline, this option should be selected. The organism and build trigger exclusion criteria and window sizes if BAIT recognizes the animal.
-8 STR
BUILD. Specifies build (eg mm9 / hg18) of species. This option will override the AS: line found in the header of the bam file. If there is no AS: line BAIT will prompt for a build unless this option is selected. Therefore, if BAIT is integrated into a pipeline, this option should be selected. The organism and build trigger exclusion criteria and window sizes if BAIT recognizes the animal.
Examples
Some test files are included in the folder test/ in the downloaded package, and can be used to confirm BAIT is correctly installed. Locate the bam file and run the following command:
BAIT -rabvB bedFileCyto -q 10 -F changeName -o testRun
The above assumes BAIT is being run in the test/ folder, otherwise you can specify the folder with -i PATH/to/test. It is possible to change the parameters for analysis, but the example given above automatically assesses SCE (-r) and plots arrowheads at their determined locations on the ideogram (-a),
together with a BED file conisting of gapped locations that is plotted on the ideograms (-G). These parameters also take only reads above a mapping quality of 20 (-q 20), automatically creates a BED file of this library (-b), and outputs everything with the file name 'testRun' (-o). Note the options -u and -U will not work for a single sample as they identify fragment regions based on shared inheritance across multiple libraries. The full dataset of mouse mm9/NCBI37 Strand-seq libraries used previously can be freely downloaded HERE from the NCBI sequence read archive under the accession number SRA055924, and converted back into bam files using the sam-dump option in the sratoolkit package.
Output Files
Files are stored in a multitude of folders in the input folder (set using -i, default is current directory). BAIT output is dependent on the options that it is fed, but will produce certain standard outputs. The following are outputs BAIT generates:
Ideogram
Options that generate output: All, except -U
File name: outputName_qualityScore_windowSize_date.pdf
Description: The standard output of a BAIT run is an ideogram of all chromosomes with histograms of reads mapping to the Watson strand (blue, left), and reads mapping to the Crick strand (orange, right). A new ideogram is plotted for each library, and contains a variety of information including the library
name, the number of reads per chromosome, the average reads per megabase, the percentage background, the coverage (if -c is used), arrowheads locating SCE regions (if -a and -r is used), and whether each chromosome is classified as WW, WC or CC. The only time this file is not plotted occurs when BAIT
is used to build a non-chromosome-stage genome when -U is invoked. The output is a pdf that is named first with an output name (default 'BAIT', set with -o option), then the quality score (default 'q10', set with the -q option), then the window size (species specific, but currently default to '200000', set with -5 option), followed by the date.
Summary File
Options that generate output: -r
Folder name: Summary_Files_outputName/
Description: The summary files generated for a BAIT run include a pdf file that incorporates a number of BAIT analyses. It plots an ideogram with lines representing SCE locations across an entire dataset (a 'composite' plot of all SCE events in all libraries) at the default binsize, and in larger bins (binsize x 5), and an ideogram of all putative mis-oriented regions. It also contains a density plot of the minimal distances between SCE events, pie charts of template strand inheritance, and a bin-by-bin plot of template inheritance across each chromosome. The folder also contains a bed-formatted SCE location file, a more complete SCE location txt le, and a table of template inheritance for each library.
BED Files
Options that generate output: -b
Folder name: BED_files_outputName
Description: Bed-format files are generated with the -b option, and are automatically formatted to be read into the UCSC genome browser. Each read is represented by a bed coordinate, and data is coerced such that Watson reads are classified as plus strand and Crick reads as minus strand, which allows strand-specific colouring using the colour-by-strand bed option. Strands are coloured identically to the ideograms, and these les can be used to manually confirm SCE events or to assess the genomic environment of any events that may have occurred. The quality score of each read is also given, and can be altered using the UCSC genome browser.
SCE locations
Options that generate output: -r
Folder name: SCE_files_outputName
Description: SCE locations determined by the DNAcopy algorithm run in BAIT are printed as both a UCSC-formatted bed file and as the states for every part of each chromosome for every Strand-seq library. Compiled files of all events are found in the summary folder. The bed files show the transition of states (such as WW->CC) in addition to the location and anueploid state. The other file shows the state on each chromosome and is used in estabilishing the inheritance state (see summary pdf) and in narrowing locations for unlocalized scaffolds if the -u option is used.
Unlocalized Scaffold Ideogram
Options that generate output: -u
Folder name: Unknown_outputName
Description: Can be thought of as aligning orphan scaffolds using the reference genome as an anchor point. Unlocalized sca old pdf has the same ideograms from the standard BAIT output, but instead of showingWatson and Crick reads, it shows percentage concordance between the templates inherited by unlocalized scaffolds and the templates inherited by chromosomes. Each page of the pdf represents a single unlocalized fragment, and displays information regarding the number of libraries used in the analysis, the percentage of WC (as low-complexity fragments will be WC in 100% of libraries), and the number of putative locations in which the fragment can map. Fragments can either be in the same orientation as the reference genome (such that a WW fragment will map to a WW chromosome) or it can be misoriented with respect to the reference (such that a WW fragment will map to a CC chromosome). As such,
histograms are plotted on the left of the ideogram for correct orientation, and on the right for mis-orientation. The region of highest concordance is shown in red on the plots. If the -G option is used, and a bed-formatted gap file is supplied, The unlocalized scaffold ideogram will assume the fragments can only reside in one of the gaps, and will output the number and locations of these regions.
Heatmap
Options that generate output: -U
File name: outputName_HEATMAP_date.pdf
Description: Can be thought of as aligning scaffolds or contigs with no reference as an anchor point. The heatmap file is generated when users are looking to construct contig- or chromosome-level genomes using the -U option. BAIT calculates the proportion of Watson and Crick reads for each fragment and creates a table of strand inheritance of each fragment in each library. This table is converted to a dissimilarity matrix, which in turn is converted to a heat map representing the similiarity of all fragments. Fragments spacially close will result in a high proportion of shared template inheritance and will cluster together. By chance, any two fragments will share the same state 50% of the time. Each
cluster should correspond to a chromosome. Note the -U option supresses all other BAIT options (as ideograms cannot be created on contig-stage genomes). A second heatmap is generated for any fragments whose template state is not WC, as these are assumed to be sex chromosomes. Subsequent plots are derived
from computationally identi ed clusters and represent subplots from the original heatmap. An ordered csv file is also generated to give the relative order of each fragment based on SCE levels.
Jump to:
Wiki Main Page
What is Strand-seq and how does it work?
Tutorial for strand inheritance studies
Tutorial for sister chromatid exchange studies
Tutorial for identifying genomic rearrangements
Tutorial for localization of orphan fragments
Tutorial for building early stage genomes

